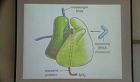
리보솜은 mRNA상의 유전정보를 단백질로 번역하는 세포에 필수적인 거대복합체이다. 이러한 리보솜은 리보핵산단백질 복합체로, rRNA와 리보솜 단백질로 이루어져있다. 리보솜 조립과정은 리...
http://chineseinput.net/에서 pinyin(병음)방식으로 중국어를 변환할 수 있습니다.
변환된 중국어를 복사하여 사용하시면 됩니다.
- 中文 을 입력하시려면 zhongwen을 입력하시고 space를누르시면됩니다.
- 北京 을 입력하시려면 beijing을 입력하시고 space를 누르시면 됩니다.
부가정보
국문 초록 (Abstract)
리보솜은 mRNA상의 유전정보를 단백질로 번역하는 세포에 필수적인 거대복합체이다. 이러한 리보솜은 리보핵산단백질 복합체로, rRNA와 리보솜 단백질로 이루어져있다. 리보솜 조립과정은 리보솜 단백질 이외에도 많은 조립인자들이 각 구성요소의 조립을 도움으로써 이루어진다. 세포 내 리보솜 조립과정에 참여하는 조립인자들로 GTPase, ATPase, 샤페론, RNA helicase, 수식효소 등 다양한 단백질들이 알려졌다. 리보솜 조립과정 중 이러한 조립인자들은 리보솜 단백질 또는 rRNA의 수식에 참여하거나, 리보솜 단백질들과 rRNA의 조립 등을 돕는다. 이러한 리보솜 조립인자들에 관한 유전학적, 구조적, 생화학적 실험결과들이 많이 존재하지만 정확한 리보솜 조립과정과 이러한 조립인자들의 역할에 대해서는 아직 밝혀지지 않았다. 현재까지의 연구결과를 바탕으로 E. coli의 리보솜 조립과정을 돕는 단백질들에 대하여 알아보고자 한다.
다국어 초록 (Multilingual Abstract)
The ribosome is a protein synthesizing machinery and a ribonucleoprotein complex that consists of three ribosomal RNAs (23S, 16S and 5S) and 54 ribosomal proteins in bacteria. In the course of ribosome assembly, ribosomal proteins (r-protein) and rRNA...
The ribosome is a protein synthesizing machinery and a ribonucleoprotein complex that consists of three ribosomal RNAs (23S, 16S and 5S) and 54 ribosomal proteins in bacteria. In the course of ribosome assembly, ribosomal proteins (r-protein) and rRNAs are modified, the r-proteins bind to rRNAs to form ribonucleoprotein complexes which are folded into mature ribosomal subunits. In this process, a number of non-ribosomal trans-acting factors organize the assembly process of the components. Those factors include GTP- and ATP-binding proteins, rRNA and r-protein modification enzymes, chaperones, and RNA helicases. During ribosome biogenesis, they participate in the modifications of ribosomal proteins and RNAs, and the assemblies of ribosomal proteins with rRNAs. Ribosomes can be assembled from a discrete set of components in vitro, and it is notable that in vivo ribosome assembly is much faster than in vitro ribosome assembly. This suggests that non-ribosomal ribosome assembly factors help to overcome several kinetic traps in ribosome biogenesis process. In spite of accumulation of genetic, structural, and biochemical data, not only the entire procedure of bacterial ribosome synthesis but also most of roles of ribosome assembly factors remain elusive. Here, we review ribosome assembly factors involved in the ribosome maturation of Escherichia coli, and summarize the contributions of several ribosome assembly factors which associate with 50S and 30S ribosomal subunits, respectively.
목차 (Table of Contents)
- 서론
- Ribosome assembly factors
- 50S ribosomal subunit assembly factors
- ObgE
- Der
- 서론
- Ribosome assembly factors
- 50S ribosomal subunit assembly factors
- ObgE
- Der
- YihI
- HflX
- DbpA
- CsdA
- SrmB
- RhlE
- RrmJ
- GroEL
- 30S ribosomal subunit assembly factors
- Era
- RsgA
- KsgA
- RbfA
- RimJ
- RimM
- RimP
- DnaK
- References
- 초록
참고문헌 (Reference)
1 Karginov, F. V., "YxiN is a modular protein combining a DEx(D/H)core and a specific RNA-binding domain" 280 : 35499-35505, 2005
2 Lindahl, L, "Two new ribosomal precursor particles in E. coli" 243 : 170-172, 1973
3 Shields, M. J., "Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli : a kinetic approach" 48 : 10793-10802, 2009
4 Nierhaus, K. H., "Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli" 71 : 4713-4717, 1974
5 Nicol, S. M., "The"DEAD box"protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA" 92 : 11681-11685, 1995
6 Engels, S., "The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum" 57 : 576-591, 2005
7 Hwang, J., "The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli" 61 : 1660-1672, 2006
8 MacDonald, R. E., "The synthesis and function of ribosomes in a new mutant of Escherichia coli" 57 : 141-147, 1967
9 Wittinghofer, A., "The structure of Ras protein: a model for a universal molecular switch" 16 : 382-387, 1991
10 Nissen, P., "The structural basis of ribosome activity in peptide bond synthesis" 289 : 920-930, 2000
1 Karginov, F. V., "YxiN is a modular protein combining a DEx(D/H)core and a specific RNA-binding domain" 280 : 35499-35505, 2005
2 Lindahl, L, "Two new ribosomal precursor particles in E. coli" 243 : 170-172, 1973
3 Shields, M. J., "Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli : a kinetic approach" 48 : 10793-10802, 2009
4 Nierhaus, K. H., "Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli" 71 : 4713-4717, 1974
5 Nicol, S. M., "The"DEAD box"protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA" 92 : 11681-11685, 1995
6 Engels, S., "The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum" 57 : 576-591, 2005
7 Hwang, J., "The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli" 61 : 1660-1672, 2006
8 MacDonald, R. E., "The synthesis and function of ribosomes in a new mutant of Escherichia coli" 57 : 141-147, 1967
9 Wittinghofer, A., "The structure of Ras protein: a model for a universal molecular switch" 16 : 382-387, 1991
10 Nissen, P., "The structural basis of ribosome activity in peptide bond synthesis" 289 : 920-930, 2000
11 Xia, B., "The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli" 332 : 575-584, 2003
12 Fischer, J. J., "The ribosome modulates the structural dynamics of the conserved GTPase HflX and triggers tight nucleotide binding" 94 : 1647-1659, 2012
13 Bystrom, A. S., "The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide" 2 : 899-905, 1983
14 Wang, S., "The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold" 12 : 959-967, 2006
15 El Hage, A., "The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli" 264 : 796-808, 2001
16 Kossen, K., "The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA" 324 : 625-636, 2002
17 Nierhaus, K. H, "The assembly of prokaryotic ribosomes" 73 : 739-755, 1991
18 Desai, P. M., "The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit" 449 : 57-63, 2006
19 Nord, S., "The RimP protein is important for maturation of the 30S ribosomal subunit" 386 : 742-753, 2009
20 Lovgren, J. M., "The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits" 10 : 1798-1812, 2004
21 Bukau, B., "The Hsp70 and Hsp60 chaperone machines" 92 : 351-366, 1998
22 Bourne, H. R., "The GTPase superfamily : a conserved switch for diverse cell functions" 348 : 125-132, 1990
23 Sato, A., "The GTP binding protein Obg homolog ObgE is involved in ribosome maturation" 10 : 393-408, 2005
24 Caldas, T., "The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase" 275 : 16414-16419, 2000
25 Bunner, A. E., "The Effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution" 398 : 1-7.9, 2010
26 Jain, C, "The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly" 14 : 381-389, 2008
27 Maki, J. A., "The DnaK chaperone system facilitates 30S ribosomal subunit assembly" 10 : 129-138, 2002
28 Charollais, J., "The DEAD‐box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli" 48 : 1253-1265, 2003
29 Inoue, K., "Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli" 48 : 1005-1016, 2003
30 Roy Chaudhuri, B., "Suppression of a cold‐sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis" 68 : 1547-1559, 2008
31 Dammel, C. S., "Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA" 9 : 626-637, 1995
32 Guthrie, C., "Studies on the assembly of ribosomes in vivo" 34 : 69-75, 1969
33 Daigle, D. M., "Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro" 186 : 1381-1387, 2004
34 Schuwirth, B. S., "Structures of the bacterial ribosome at 3. 5 A resolution" 310 : 827-834, 2005
35 Selmer, M., "Structure of the 70S ribosome complexed with mRNA and tRNA" 313 : 1935-1942, 2006
36 Tu, C., "Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis" 106 : 14843-14848, 2009
37 Traub, P., "Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins" 59 : 777-784, 1968
38 Guthrie, C., "Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly" 63 : 384-391, 1969
39 Boehringer, D., "Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis" 287 : 10453-10459, 2012
40 Suzuki, S., "Structural characterization of the ribosome maturation protein, RimM" 189 : 6397-6406, 2007
41 Guo, Q., "Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy" 108 : 13100-13105, 2011
42 Datta, P. P., "Structural aspects of RbfA action during small ribosomal subunit assembly" 28 : 434-445, 2007
43 Feng, B., "Structural and functional insights into the mode of action of a universally conserved Obg GTPase" 12 : e1001866-, 2014
44 Trubetskoy, D., "SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo" 37 : 6540-6549, 2009
45 Inoue, K., "Specific growth inhibition by acetate of an Escherichia coli strain expressing Era-dE, a dominant negative Era mutant" 4 : 379-388, 2002
46 Nishimura, M., "Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8" 344 : 1369-1383, 2004
47 Morimoto, T., "Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis" 148 : 3539-3552, 2002
48 Goto, S., "RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis" 30 : 104-114, 2011
49 Britton, R. A, "Role of GTPases in bacterial ribosome assembly" 63 : 155-176, 2009
50 Bylund, G. O., "RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli" 180 : 73-82, 1998
51 Kimura, T., "Ribosome-small-subunit-dependent GTPase interacts with tRNA-binding sites on the ribosome" 381 : 467-477, 2008
52 Peil, L., "Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA" 275 : 3772-3782, 2008
53 Niereaus, K. H., "Ribosomal proteins" 74 : 587-597, 1973
54 Hase, Y., "Removal of a ribosome small subunit-dependent GTPase confers salt resistance on Escherichia coli cells" 15 : 1766-1774, 2009
55 Khaitovich, P., "Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA" 38 : 1780-1788, 1999
56 Green, R., "Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA" 38 : 1772-1779, 1999
57 O'Farrell, H. C., "Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution" 12 : 725-733, 2006
58 Jones, P. G., "RbfA, a 30S ribosomal binding factor, is a cold‐shock protein whose absence triggers the cold‐shock response" 21 : 1207-1218, 1996
59 Bügl, H., "RNA methylation under heat shock control" 6 : 349-360, 2000
60 Dutta, D., "Properties of HflX, an enigmatic protein from Escherichia coli" 191 : 2307-2314, 2009
61 Prud’homme-Généreux, A., "Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold‐shock protein, CsdA : evidence for a ‘cold shock degradosome’" 54 : 1409-1421, 2004
62 Tan, J., "Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase" 2692-2698, 2002
63 Connolly, K., "Overexpression of RbfA in the absence of the KsgA checkpoint results in impaired translation initiation" 87 : 968-981, 2013
64 Elles, L. M., "Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype" 36 : 41-50, 2008
65 Alix, J. H., "Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli" 90 : 9725-9729, 1993
66 Thammana, P, "Methylation of 16S RNA during ribosome assembly in vitro" 251 : 682-686, 1974
67 Connolly, K., "Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA" 70 : 1062-1075, 2008
68 Helser, T. L., "Mechanism of kasugamycin resistance in Escherichia coli" 235 : 6-9, 1972
69 McCutcheon, J. P., "Location of translational initiation factor IF3 on the small ribosomal subunit" 96 : 4301-4306, 1999
70 Tsu, C. A., "Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA" 37 : 16989-16996, 1998
71 Lindahl, L, "Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes" 92 : 15-37, 1975
72 Karginov, F. V., "Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme" 32 : 3028-3032, 2004
73 Sharma, M. R., "Interaction of Era with the 30S ribosomal subunit : implications for 30S subunit assembly" 18 : 319-329, 2005
74 Krzyzosiak, W., "In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome" 26 : 2353-2364, 1987
75 Houry, W. A., "Identification of in vivo substrates of the chaperonin GroEL" 402 : 147-154, 1999
76 Koller-Eichhorn, R., "Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins" 282 : 19928-19937, 2007
77 Sohlberg, B., "Functional interaction of heat shock protein GroEL with an RNase E-like activity in Escherichia coli" 90 : 277-281, 1993
78 Diges, C. M., "Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA" 2 : 5503-5512, 2001
79 Sayed, A., "Era, an Essential Escherichia coli Small G-Protein, Binds to the 30S Ribosomal Subunit" 264 : 51-54, 1999
80 Meier, T. I., "Era GTPase of Escherichia coli : binding to 16S rRNA and modulation of GTPase activity by RNA and carbohydrates" 146 (146): 1071-1083, 2000
81 Moll, I., "Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD‐box helicase on translation of leaderless and canonical mRNAs in Escherichia coli" 44 : 1387-1396, 2002
82 Jain, N., "E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides" 379 : 201-205, 2009
83 Tomar, S. K., "Distinct GDP/GTP bound states of the tandem G-domains of EngA regulate ribosome binding" 37 : 2359-2370, 2009
84 Guo, Q., "Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process" 41 : 2609-2620, 2013
85 Iost, I., "DEAD-box RNA helicases in Escherichia coli" 34 : 4189-4197, 2006
86 Charollais, J., "CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit" 32 : 2751-2759, 2004
87 Carter, A. P., "Crystal structure of an initiation factor bound to the 30S ribosomal subunit" 291 : 498-501, 2001
88 Chen, X., "Crystal structure of ERA : a GTPase-dependent cell cycle regulator containing an RNA binding motif" 96 : 8396-8401, 1999
89 Pillutla, R. C., "Cross-species complementation of the indispensable Escherichia coli era gene highlights amino acid regions essential for activity" 177 : 2194-2196, 1995
90 Polach, K. J., "Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA" 41 : 3693-3702, 2002
91 Bharat, A., "Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA" 188 : 7992-7996, 2006
92 Lerner, C. G., "Cold‐sensitive conditional mutations in Era, an essential Escherichia coli GTPase, isolated by localized random polymerase chain reaction mutagenesis" 126 : 291-298, 1995
93 Jones, P. G., "Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli" 93 : 76-80, 1996
94 Thieringer, H. A., "Cold shock and adaptation" 20 : 49-57, 1998
95 Yoshikawa, A., "Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12" 209 : 481-488, 1987
96 Boddeker, N., "Characterization of DbpA, an Escherichia coli DEAD box protein with ATP independent RNA unwinding activity" 25 : 537-545, 1997
97 Britton, R. A., "Cell cycle arrest in Era GTPase mutants : a potential growth rate‐regulated checkpoint in Escherichia coli" 27 : 739-750, 1998
98 Hayes, F., "Biosynthesis of ribosomes in E. coli : I. —Properties of ribosomal precursor particles and their RNA components" 53 : 369-382, 1971
99 Hage, A. E., "Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone" 51 : 189-201, 2004
100 Williamson, J. R, "Assembly of the 30S ribosomal subunit" 38 : 397-403, 2005
101 Spillmann, S., "Assembly in Vitro of the 50 S subunit from Escherichia coli ribosomes : Proteins essential for the first heat-dependent conformational change" 115 : 513-523, 1977
102 Holmes, K. L., "Analysis of conformational changes in 16S rRNA during the course of 30S subunit assembly" 354 : 340-357, 2005
103 Hwang, J., "An essential GTPase, der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima" 276 : 31415-31421, 2001
104 Nishi, K., "An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly" 336 : 496-498, 1988
105 Lewandowski, L. J., "An altered pattern of ribosome synthesis in a mutant of E. coli" 25 : 554-561, 1966
106 Blombach, F., "An HflX-type GTPase from Sulfolobus solfataricus binds to the 50S ribosomal subunit in all nucleotide-bound states" 193 : 2861-2867, 2011
107 Williamson, J. R, "After the ribosome structures: how are the subunits assembled?" 9 : 165-167, 2003
108 Hager, J., "Active site in RrmJ, a heat shock-induced methyltransferase" 277 : 41978-41986, 2002
109 Kirthi, N., "A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity" 12 : 2080-2091, 2006
110 Bylund, G. O., "A novel ribosome-associated protein is important for efficient translation in Escherichia coli" 179 : 4567-4574, 1997
111 Himeno, H., "A novel GTPase activated by the small subunit of ribosome" 32 : 5303-5309, 2004
112 Sharpe Elles, L. M., "A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit" 37 : 6503-6514, 2009
113 Xu, Z., "A conserved rRNA methyltransferase regulates ribosome biogenesis" 15 : 534-536, 2008
114 Hwang, J., "A Bacterial GAP-Like Protein, YihI, Regulating the GTPase of Der, an Essential GTP-Binding Protein in Escherichia coli" 399 : 759-772, 2010
동일학술지(권/호) 다른 논문
-
Spatial Distribution Pattern of the Populations of Camellia japonica in Busan
- 한국생명과학회
- Man Ki Kang(강만기)
- 2014
- KCI등재
-
Parkin Interacts with the PDZ Domain of Multi-PDZ Domain Protein MUPP1
- 한국생명과학회
- Won Hee Jang(장원희)
- 2014
- KCI등재
-
Aspergillus oryzae로 생물전환한 감귤박의 항비만 효과
- 한국생명과학회
- 전현주(Hyun Joo Jeon)
- 2014
- KCI등재
-
사람 폐 섬유아 세포에서 Brunfelsia grandiflora 에탄올 추출물이 Autophagy에 미치는 영향
- 한국생명과학회
- 남향(Hyang Nam)
- 2014
- KCI등재
분석정보
인용정보 인용지수 설명보기
학술지 이력
| 연월일 | 이력구분 | 이력상세 | 등재구분 |
|---|---|---|---|
| 2027 | 평가예정 | 재인증평가 신청대상 (재인증) | |
| 2021-01-01 | 평가 | 등재학술지 유지 (재인증) |  |
| 2018-01-01 | 평가 | 등재학술지 유지 (등재유지) |  |
| 2015-01-01 | 평가 | 등재학술지 유지 (등재유지) |  |
| 2011-08-03 | 학술지명변경 | 외국어명 : Korean Journal of Life Science -> Journal of Life Science |  |
| 2011-01-01 | 평가 | 등재학술지 유지 (등재유지) |  |
| 2009-01-01 | 평가 | 등재학술지 유지 (등재유지) |  |
| 2007-01-01 | 평가 | 등재학술지 유지 (등재유지) |  |
| 2004-01-01 | 평가 | 등재학술지 선정 (등재후보2차) |  |
| 2003-01-01 | 평가 | 등재후보 1차 PASS (등재후보1차) |  |
| 2001-07-01 | 평가 | 등재후보학술지 선정 (신규평가) |  |
학술지 인용정보
| 기준연도 | WOS-KCI 통합IF(2년) | KCIF(2년) | KCIF(3년) |
|---|---|---|---|
| 2016 | 0.37 | 0.37 | 0.42 |
| KCIF(4년) | KCIF(5년) | 중심성지수(3년) | 즉시성지수 |
| 0.43 | 0.43 | 0.774 | 0.09 |





 ScienceON
ScienceON DBpia
DBpia